Neuronal morphology analysis and classification¶
All scripts used for the analysis of the neuron segmentation are located in SyConn/scripts/multiviews_neuron/, namely:
SSV multi-views generation:
start_sso_rendering.pyCell compartment prediction:
axoness_prediction.pyandspiness_prediction.pyCell type prediction:
celltype_prediction.py
Prerequisites¶
Knossos- and SegmentationDataset of the supervoxel segmentation
SegmentationDatasets for all cellular organelles (currently mitochondria, vesicle clouds and synaptic junctions)
[Optional] Glia removal
The RAG/SV-mapping in form of a SSD, see SuperSegmentationDatasets
Mapped cellular organelles to SSVs
Steps¶
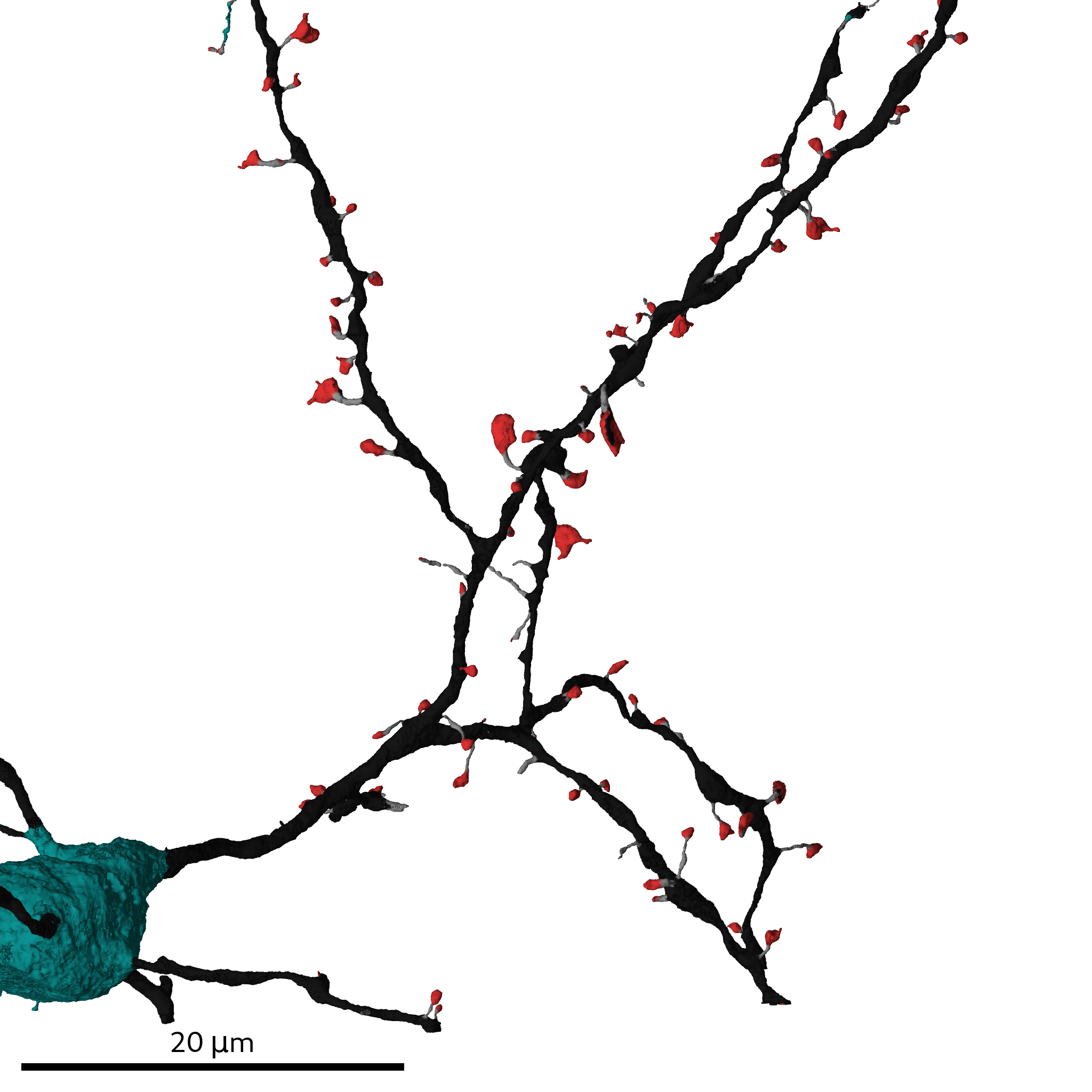
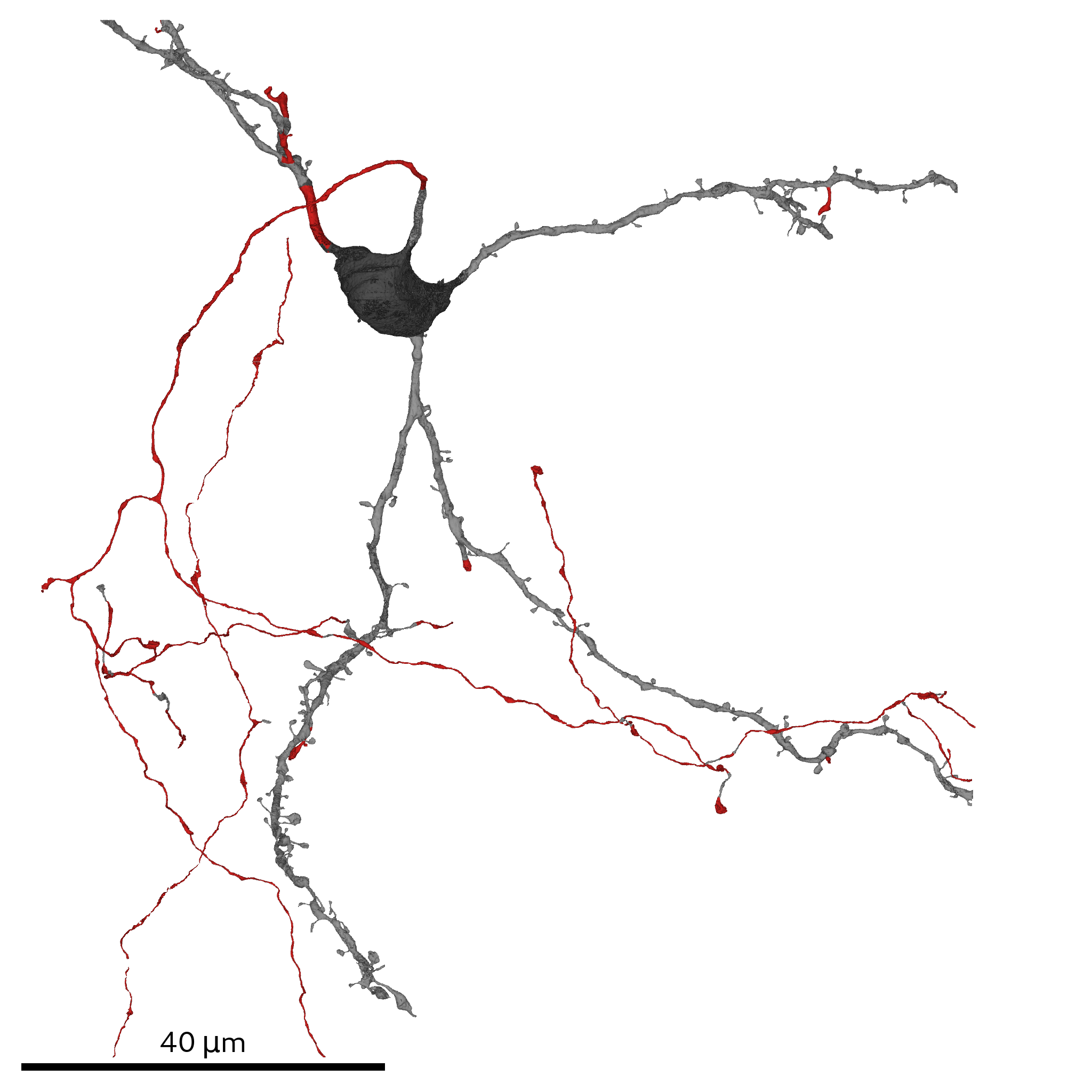 The multi-views contain shape information of cellular organelles and the cell/SSV outline
and are the basis for predicting cell compartments, cell type and spines. To generate them dataset-wide run:
The multi-views contain shape information of cellular organelles and the cell/SSV outline
and are the basis for predicting cell compartments, cell type and spines. To generate them dataset-wide run:
start_sso_rendering.py
Dataset-wide predictions can be started via the respective scripts:
Cell compartments:
axoness_prediction.pyandspiness_prediction.pyCell types:
celltype_prediction.py
Their results are stored on SSV level and used for extracting the wiring diagram later on.