Glia removal¶
All scripts used for the analysis of the neuron segmentation are located in SyConn/scripts/multiviews_glia/.
Prerequisites¶
Knossos- and SegmentationDataset of the supervoxel segmentation
Initial RAG/SV-mapping
Steps¶
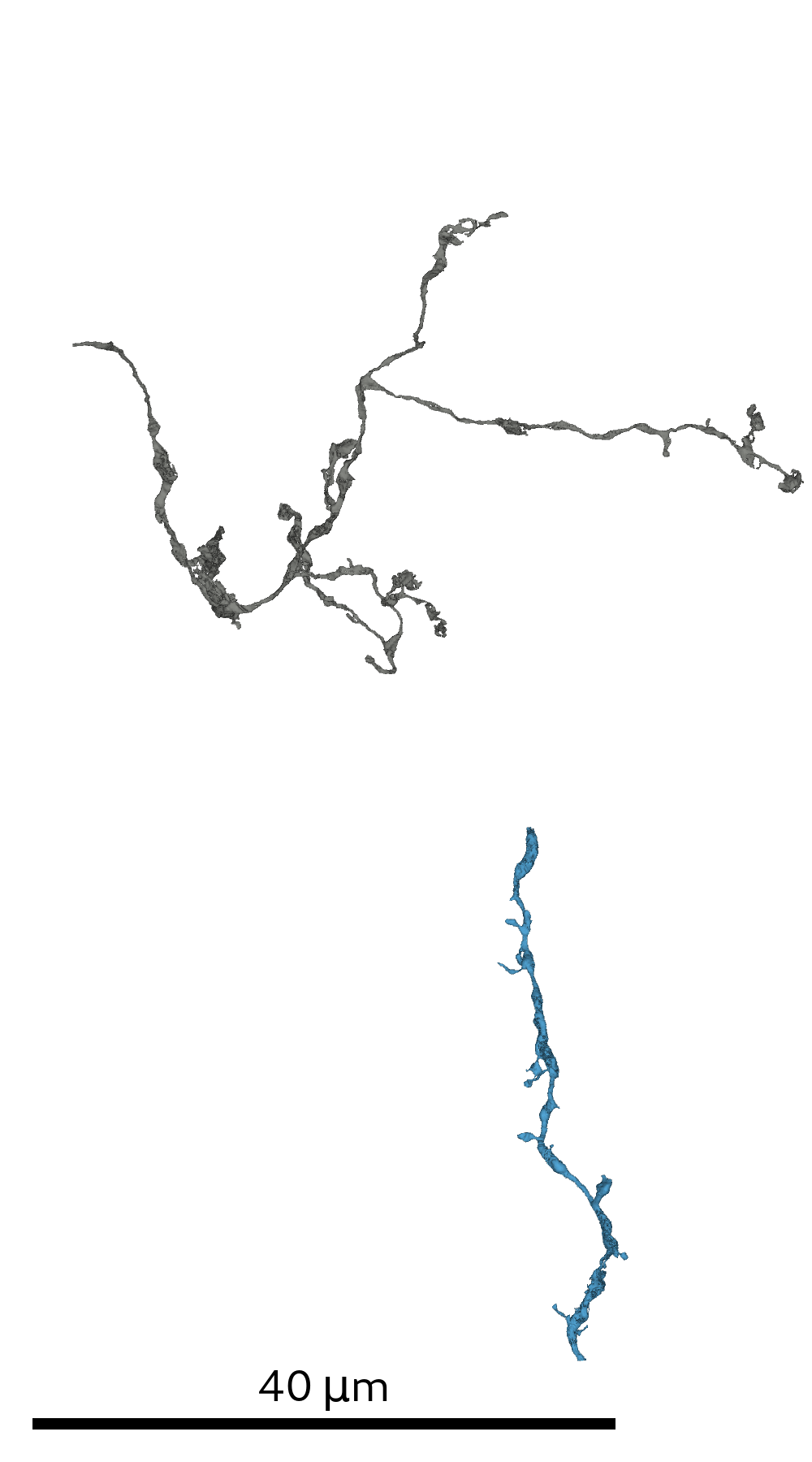
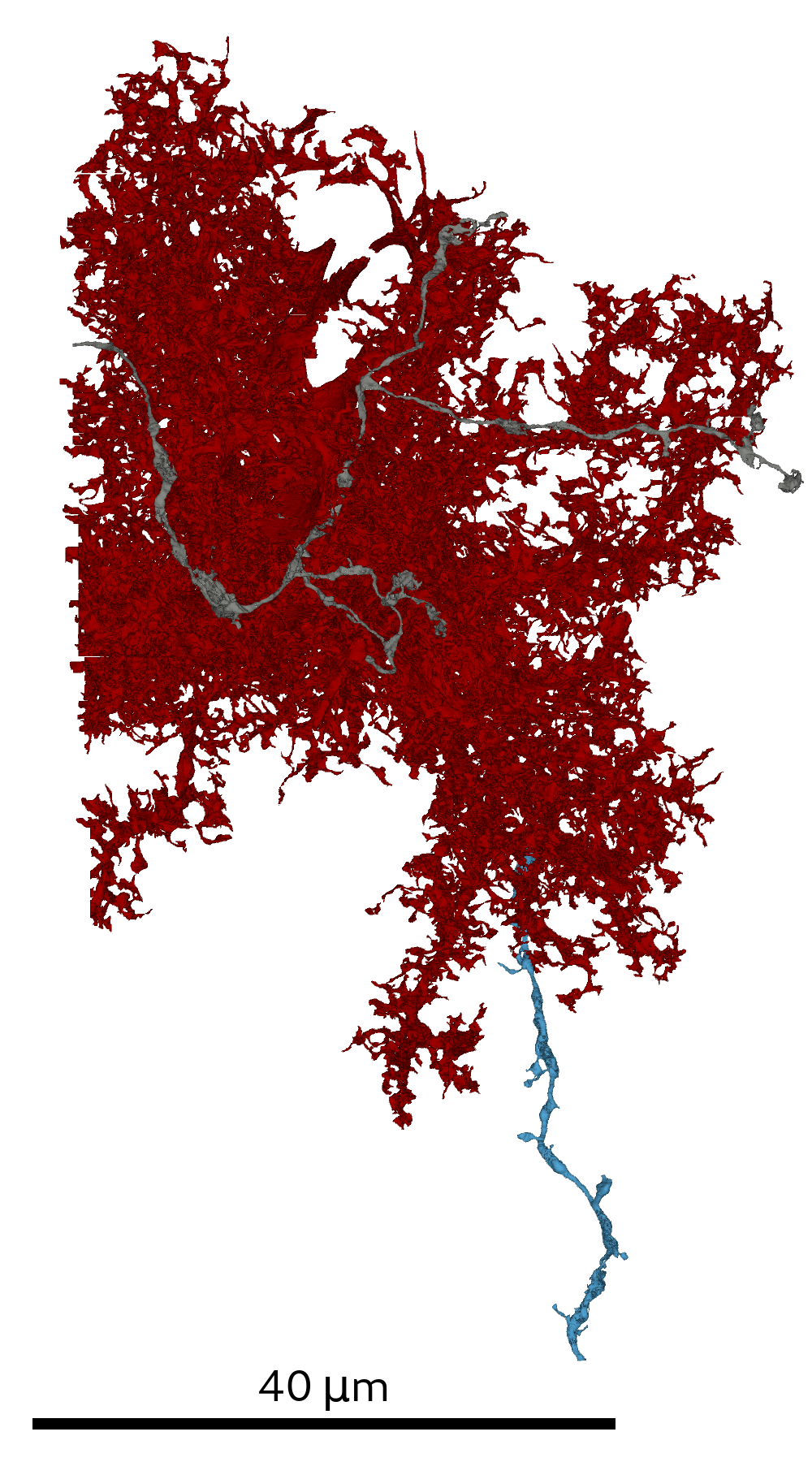 In order to generate the SSV multi-views prior to glia removal it is necessary to provide the script with an initial RAG/SV-mapping.
The agglomerated SVs are required to provide sufficient context for the glia prediction:
In order to generate the SSV multi-views prior to glia removal it is necessary to provide the script with an initial RAG/SV-mapping.
The agglomerated SVs are required to provide sufficient context for the glia prediction:
start_sso_rendering_glia_removal.py
In order to start the glia prediction run:
glia_prediction.py
SVs with predicted glia labels will be removed via a splitting heuristic. For splitting and generating the glia-free region adjacency graph (RAG) run:
glia_splitting.py